lördag 20 maj 2023
Amalgamaa ajatellen
Metallotioniinien geeniperheestä MTn GeneCards luettelosta: MT1, MT2, MT3 ja MT4 . Osa on klusterina.
https://www.genecards.org/cgi-bin/carddisp.pl?gene=MT1A&keywords=metallothionein
*MT1A
-
This gene is a member of the metallothionein family of genes. Proteins encoded by this gene family are low in molecular weight, are cysteine-rich, lack aromatic residues, and bind divalent heavy metal ions. The conserved cysteine residues co-ordinate metal ions using mercaptide linkages. These proteins act as anti-oxidants, protect against hydroxyl free radicals, are important in homeostatic control of metal in the cell, and play a role in detoxification of heavy metals. Disruption of two metallothionein genes in mouse resulted in defects in protection against heavy metals, oxidative stress, immune reactions, carcinogens, and displayed obesity. [provided by RefSeq, Sep 2017]
GeneCards Summary for MT1A Gene
MT1A (Metallothionein 1A) is a Protein Coding gene. Diseases associated with MT1A include Menkes Disease and Deficiency Anemia. Among its related pathways are Metal ion SLC transporters and Cellular responses to stimuli. An important paralog of this gene is MT2A.
UniProtKB/Swiss-Prot Summary for MT1A Gene
Metallothioneins have a high content of cysteine residues that bind various heavy metals; these proteins are transcriptionally regulated by both heavy metals and glucocorticoids. ( MT1A_HUMAN,P04731 )
* MT1B
-
The protein encoded by this gene binds heavy metals and protects against toxicity from heavy metal ions. This gene is found in a cluster of similar genes on chromosome 16. [provided by RefSeq, Jul 2016]
GeneCards Summary for MT1B Gene
MT1B (Metallothionein 1B) is a Protein Coding gene. Diseases associated with MT1B include Deficiency Anemia. Among its related pathways are Metal ion SLC transporters and Cellular responses to stimuli. An important paralog of this gene is MT1X.
UniProtKB/Swiss-Prot Summary for MT1B Gene
Metallothioneins have a high content of cysteine residues that bind various heavy metals; these proteins are transcriptionally regulated by both heavy metals and glucocorticoids. ( MT1B_HUMAN,P07438 )
* MTC1P, pseudogene
https://www.genecards.org/cgi-bin/carddisp.pl?gene=MT1CP&keywords=MT1F
Molecular function for MT1CP Gene according to GENATLAS
- Biochemistry:
-
- metallothionein 1CP cluster MT1CP
* MT1DP pseudogene
https://www.genecards.org/cgi-bin/carddisp.pl?gene=MT1DP&keywords=MT1F
-
Predicted to enable metal ion binding activity. Predicted to be involved in cellular response to metal ion; cellular zinc ion homeostasis; and detoxification of copper ion. Predicted to be active in cytoplasm and nucleus. [provided by Alliance of Genome Resources, Apr 2022]
GeneCards Summary for MT1DP Gene
MT1DP (Metallothionein 1D, Pseudogene) is a Pseudogene. Diseases associated with MT1DP include Gastric Cancer and Hepatocellular Carcinoma.
UniProtKB/Swiss-Prot Summary for MT1DP Gene
Metallothioneins have a high content of cysteine residues that bind various heavy metals. ( MT1DP_HUMAN,A1L3X4 )
* MT1E
-
Predicted to enable zinc ion binding activity. Involved in cellular response to cadmium ion and cellular response to zinc ion. Located in cytoplasm and nucleus. [provided by Alliance of Genome Resources, Apr 2022]
GeneCards Summary for MT1E Gene
MT1E (Metallothionein 1E) is a Protein Coding gene. Diseases associated with MT1E include Frontometaphyseal Dysplasia 1 and Bladder Cancer. Among its related pathways are Metal ion SLC transporters and Cellular responses to stimuli. An important paralog of this gene is MT2A.
* MT1F
-
Predicted to enable zinc ion binding activity. Involved in cellular response to cadmium ion. Located in cytoplasm and nucleus. [provided by Alliance of Genome Resources, Apr 2022]
GeneCards Summary for MT1F Gene
MT1F (Metallothionein 1F) is a Protein Coding gene.
Diseases associated with MT1F include Deficiency Anemia.
Among its related pathways are Metal ion SLC transporters and Cellular responses to stimuli.
An important paralog of this gene is MT1G.
*MT1G
-
Enables zinc ion binding activity. Involved in cellular response to metal ion; cellular response to vascular endothelial growth factor stimulus; and negative regulation of growth. Located in cytoplasm and nucleus. [provided by Alliance of Genome Resources, Apr 2022]
GeneCards Summary for MT1G Gene
MT1G (Metallothionein 1G) is a Protein Coding gene. Diseases associated with MT1G include Hepatocellular Carcinoma and Deficiency Anemia. Among its related pathways are Metal ion SLC transporters and Cellular responses to stimuli. An important paralog of this gene is MT2A.
* MT1H
-
Predicted to enable zinc ion binding activity. Involved in cellular response to cadmium ion and cellular response to zinc ion. Predicted to be active in cytoplasm and nucleus. [provided by Alliance of Genome Resources, Apr 2022]
GeneCards Summary for MT1H Gene
MT1H (Metallothionein 1H) is a Protein Coding gene. Diseases associated with MT1H include Deficiency Anemia. Among its related pathways are Metal ion SLC transporters and Cellular responses to stimuli. An important paralog of this gene is MT1HL1.
*MT1L
-
Predicted to enable zinc ion binding activity. Involved in cellular response to zinc ion. Predicted to be active in cytoplasm and nucleus. [provided by Alliance of Genome Resources, Apr 2022]
GeneCards Summary for MT1L Gene
MT1L (Metallothionein 1L, Pseudogene) is a Pseudogene. Among its related pathways are Metal ion SLC transporters and Copper homeostasis.
UniProtKB/Swiss-Prot Summary for MT1L Gene
Metallothioneins have a high content of cysteine residues that bind various heavy metals; these proteins are transcriptionally regulated by both heavy metals and glucocorticoids. ( MT1L_HUMAN,Q93083 )
* MT1M ( alias * MT1K)
-
This gene encodes a member of the metallothionein superfamily, type 1 family. Metallothioneins have a high content of cysteine residues that bind various heavy metals. These genes are transcriptionally regulated by both heavy metals and glucocorticoids. [provided by RefSeq, Oct 2011]
GeneCards Summary for MT1M Gene
MT1M (Metallothionein 1M) is a Protein Coding gene. Diseases associated with MT1M include Deficiency Anemia. Among its related pathways are Metal ion SLC transporters and Cellular responses to stimuli. An important paralog of this gene is MT1B.
UniProtKB/Swiss-Prot Summary for MT1M Gene
Metallothioneins have a high content of cysteine residues that bind various heavy metals; these proteins are transcriptionally regulated by both heavy metals and glucocorticoids. ( MT1M_HUMAN,Q8N339 )
*MT1X
-
Predicted to enable copper ion binding activity and zinc ion binding activity. Involved in cellular response to cadmium ion; cellular response to erythropoietin; and cellular response to zinc ion. Located in cytoplasm and nucleus. [provided by Alliance of Genome Resources, Apr 2022]
GeneCards Summary for MT1X Gene
MT1X (Metallothionein 1X) is a Protein Coding gene. Diseases associated with MT1X include Deficiency Anemia. Among its related pathways are Metal ion SLC transporters and Cellular responses to stimuli. An important paralog of this gene is MT2A.
UniProtKB/Swiss-Prot Summary for MT1X Gene
Metallothioneins have a high content of cysteine residues that bind various heavy metals; these proteins are transcriptionally regulated by both heavy metals and glucocorticoids. May be involved in FAM168A anti-apoptotic signaling (PubMed:23251525). ( MT1X_HUMAN,P80297 )
*MT2
https://www.genecards.org/cgi-bin/carddisp.pl?gene=MT2A&keywords=MT2
-
This gene is a member of the metallothionein family of genes. Proteins encoded by this gene family are low in molecular weight, are cysteine-rich, lack aromatic residues, and bind divalent heavy metal ions, altering the intracellular concentration of heavy metals in the cell. These proteins act as anti-oxidants, protect against hydroxyl free radicals, are important in homeostatic control of metal in the cell, and play a role in detoxification of heavy metals. The encoded protein interacts with the protein encoded by the homeobox containing 1 gene in some cell types, controlling intracellular zinc levels, affecting apoptotic and autophagy pathways. Some polymorphisms in this gene are associated with an increased risk of cancer. [provided by RefSeq, Sep 2017]
GeneCards Summary for MT2A Gene
MT2A (Metallothionein 2A) is a Protein Coding gene. Diseases associated with MT2A include Scrapie and Osteogenesis Imperfecta, Type X. Among its related pathways are Metal ion SLC transporters and Interferon gamma signaling. Gene Ontology (GO) annotations related to this gene include obsolete drug binding. An important paralog of this gene is MT1G.
UniProtKB/Swiss-Prot Summary for MT2A Gene
Metallothioneins have a high content of cysteine residues that bind various heavy metals; these proteins are transcriptionally regulated by both heavy metals and glucocorticoids. ( MT2_HUMAN,P02795 )
* MT3
https://www.genecards.org/cgi-bin/carddisp.pl?gene=MT3&keywords=MT3
-
This gene is a member of the metallothionein family of genes. Proteins encoded by this gene family are low in molecular weight, are cysteine-rich, lack aromatic residues, and bind divalent heavy metal ions. This gene family member displays tissue-specific expression, and contains a threonine insert near its N-terminus and a glutamate-rich hexapeptide insert near its C-terminus relative to the proteins encoded by other gene family members. It plays an important role in zinc and copper homeostasis, and is induced under hypoxic conditions. The encoded protein is a growth inhibitory factor, and reduced levels of the protein are observed in the brains of individuals with some metal-linked neurodegenerative disorders such as Alzheimer's disease. [provided by RefSeq, Sep 2017]
*MT4
https://www.genecards.org/cgi-bin/carddisp.pl?gene=MT4&keywords=MT3
-
Predicted to enable metal ion binding activity. Predicted to be involved in cellular response to metal ion; cellular zinc ion homeostasis; and detoxification of copper ion. Predicted to act upstream of or within cellular metal ion homeostasis. Predicted to be active in cytoplasm and nucleus. [provided by Alliance of Genome Resources, Apr 2022]
MetO, sulfoxidimetioniinia kuvataan Wikipediasivulla näin:

| |
| Names | |
|---|---|
| IUPAC name
2-Amino-4-(methylsulfinyl)butanoic acid
| |
| Identifiers | |
| |
3D model (JSmol)
|
|
| ChemSpider |
|
| ECHA InfoCard | 100.057.891 |
| EC Number |
|
PubChem CID
|
|
| UNII |
|
CompTox Dashboard (EPA)
|
|
| Properties | |
|---|---|
| C5H11NO3S | |
| Molar mass | 165.21 g·mol−1 |
| Appearance | white solid |
| Hazards | |
| GHS labelling: | |

| |
| Warning | |
| H315, H319, H335 | |
| P261, P264, P271, P280, P302+P352, P304+P340, P305+P351+P338, P312, P321, P332+P313, P337+P313, P362, P403+P233, P405, P501 | |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
| |
Methionine sulfoxide is the organic compound with the formula CH3S(O)CH2CH2CH(NH2)CO2H. It is an amino acid that occurs naturally although it is formed post-translationally (PTM).
Oxidation of the sulfur of methionine results in methionine sulfoxide or methionine sulfone. The sulfur-containing amino acids methionine and cysteine are more easily oxidized than the other amino acids.[1][2] Unlike oxidation of other amino acids, the oxidation of methionine can be reversed by enzymatic action, specifically by enzymes in the methionine sulfoxide reductase family of enzymes. The three known methionine sulfoxide reductases are MsrA, MsrB, and fRmsr.[2] Oxidation of methionine results in a mixture of the two diastereomers methionine-S-sulfoxide and methionine-R-sulfoxide, which are reduced by MsrA and MsrB, respectively.[3] MsrA can reduce both free and protein-based methionine-S-sulfoxide, whereas MsrB is specific for protein-based methionine-R-sulfoxide. fRmsr, however, catalyzes the reduction of free methionine-R-sulfoxide.[2] Thioredoxin serves to recycle by reduction some of the methionine sulfoxide reductase family of enzymes, whereas others can be reduced by metallothionein.[4] Biochemical function
Methionine sulfoxide (MetO), the oxidized form of the amino acid methionine (Met), increases with age in body tissues, which is believed by some to contribute to biological ageing.[5][6] Oxidation of methionine residues in tissue proteins can cause them to misfold or otherwise render them dysfunctional.[5] Uniquely, the methionine sulfoxide reductase (Msr) group of enzymes act with thioredoxin to catalyze the enzymatic reduction and repair of oxidized methionine residues.[5] Moreover, levels of methionine sulfoxide reductase A (MsrA) decline in aging tissues in mice and in association with age-related disease in humans.[5] There is thus a rationale for thinking that by maintaining the structure, increased levels or activity of MsrA might retard the rate of aging.
Indeed, transgenic Drosophila (fruit flies) that overexpress methionine sulfoxide reductase show extended lifespan.[7] However, the effects of MsrA overexpression in mice were ambiguous.[8] MsrA is found in both the cytosol and the energy-producing mitochondria, where most of the body's endogenous free radicals are produced. Transgenically increasing the levels of MsrA in either the cytosol or the mitochondria had no significant effect on lifespan assessed by most standard statistical tests, and may possibly have led to early deaths in the cytosol-specific mice, although the survival curves appeared to suggest a slight increase in maximum (90%) survivorship, as did analysis using Boschloo's Exact test, a binomial test designed to test greater extreme variation.[8]
The oxidation of methionine serves as a switch that deactivates certain protein activities such as E.coli ribosomal protein, L12.[9] Proteins with great amount of methionine residues tend to exist within the lipid bilayer as methionine is one of the most hydrophobic amino acids. Those methionine residues that are exposed to the aqueous exterior thus are vulnerable to oxidation. The oxidized residues tend to be arrayed around the active site and may guard access to this site by reactive oxygen species. Once oxidized, the MetO residues are reduced back to methionine by the enzyme methionine sulfoxide reductase. Thus, an oxidation–reduction cycle occurs in which exposed methionine residues are oxidized (e.g., by H2O2) to methionine sulfoxide residues, which are subsequently reduced.[10]
Methionine(protein)+ H2O2→ Methionine Sulfoxide(protein)+ H2O
Methionine Sulfoxide(protein)+ NADPH+H+→ Methionine(protein)+ NADP++H2O
Essentielli aminohappo metioniini (Met, M) palautuu reduktaaseilla sulfoxidimuodoistaan.
MSRA (8p23.1), 235 a.a.. "Mithochondrial peptide methionine sulfoxide reductase".
https://www.genecards.org/cgi-bin/carddisp.pl?gene=MSRA&keywords=Methionine-S-sulfoxide,reductase;
- Mitochondrial Peptide Methionine Sulfoxide Reductase 3 4
- Peptide-Methionine (S)-S-Oxide Reductase 3 4
- Peptide Met(O) Reductase 3 4
This gene encodes a ubiquitous and highly conserved protein that carries out the enzymatic reduction of methionine sulfoxide to methionine. Human and animal studies have shown the highest levels of expression in kidney and nervous tissue. The protein functions in the repair of oxidatively damaged proteins to restore biological activity. Alternative splicing results in multiple transcript variants. [provided by RefSeq, May 2014]
MSRB1 (16p13.3), 116 a.a. "Methionine-R-sulfoxide reductase B1"
Cofactor Zn++.
https://www.genecards.org/cgi-bin/carddisp.pl?gene=MSRB1&keywords=Methionine-S-sulfoxide,reductase;
The protein encoded by this gene belongs to the methionine-R-sulfoxide reductase B (MsrB) family. Members of this family function as repair enzymes that protect proteins from oxidative stress by catalyzing the reduction of methionine-R-sulfoxides to methionines. This protein is highly expressed in liver and kidney, and is localized to the nucleus and cytosol. It is the only member of the MsrB family that is a selenoprotein, containing a selenocysteine (Sec) residue at its active site. It also has the highest methionine-R-sulfoxide reductase activity compared to other members containing cysteine in place of Sec. Sec is encoded by the UGA codon, which normally signals translation termination. The 3' UTRs of selenoprotein mRNAs contain a conserved stem-loop structure, designated the Sec insertion sequence (SECIS) element, that is necessary for the recognition of UGA as a Sec codon, rather than as a stop signal. A pseudogene of this locus has been identified on chromosome 19. [provided by RefSeq, Aug 2017]
Methionine-sulfoxide reductase that specifically reduces methionine (R)-sulfoxide back to methionine. While in many cases, methionine oxidation is the result of random oxidation following oxidative stress, methionine oxidation is also a post-translational modification that takes place on specific residue. Acts as a regulator of actin assembly by reducing methionine (R)-sulfoxide mediated by MICALs (MICAL1, MICAL2 or MICAL3) on actin, thereby promoting filament repolymerization. Plays a role in innate immunity by reducing oxidized actin, leading to actin repolymerization in macrophages.
MSRB2, (10p12.2), 182 a.a., " Methionine-(R)- sulfoxide reductase B2, mithochondrial".
MSRB3, (12q14.3) ,192 a.a., "Methionine R-Sulfoxide reductase B3"
https://www.genecards.org/cgi-bin/carddisp.pl?gene=MSRB3
https://www.genecards.org/cgi-bin/carddisp.pl?gene=MSRB2&keywords=Methionine-S-sulfoxide,reductase; (Tästä on löydetty aktiivisuutta Stafylokokki aureusta vastaan)
fredag 12 maj 2023
TAURIINI ja hypotauriini. Retinan tila Artikkeli pohdittavaksi.
https://www.mdpi.com/2218-1989/9/7/127
SAH, S-adenosyl-homocystein sah
Tämän aminohapon ominaisarvosta HAKU:
SAH on jäljellä kun SAM (Aktivoitu metioniini, S-adenosylmetioniini) on luovuttanut metyyliryhmänsä (Ch3-) substraatille.
Metioniinia voi palautua mutta SAH muto omaa muitakin teitä.
Mitä ne ovat? Mikä merkitys on tällä välituotteella SAH?
Etsin artikkeleita, joissa se mainitaan SAH
(1)
Targeting Histone Methylation
Marco P. Licciardello, Stefan Kubicek, in Drug Discovery in Cancer Epigenetics, 2016
Ten Ways to Target Histone Methylation 219
- 9.3.1 Methyltransferases can be Inhibited by Preventing SAH Turnover 219
- 9.3.2 Targeting the Peptide-Binding Pocket of Methyltransferases 221
- 9.3.3 The SAM-Binding Pocket as Universal Target of all Methyltransferases 222
- 9.3.4 Allosteric and Indirect Inhibition of Methyltransferases 222
- 9.3.5 Irreversible Inhibitors of LSD1/2 Covalently Bind the Cofactor FAD 222
- 9.3.6 The Emergence of the First Substrate Competitive LSD1 Inhibitors 224
- 9.3.7 Metabolic Targeting of JmjC Demethylases 224
- 9.3.8 Potent Small-Molecule Inhibitors Binding the Active Sites of JmjC Demethylases 224
- 9.3.9 PRMTs and PADs 225
- 9.3.10 Methylation Modulators of the Future 225
- (2)
https://doi.org/10.1016/S0065-3527(03)61014-6
Prospects for Antiviral Therapy The Flaviviruses: Detection, Diagnosis, and Vaccine Development
P. Leyssen, ... J. Neyts, in Advances in Virus Research, 2003
S-adenosylhomocysteine Hydrolase
The SAH hydrolase is a pivotal enzyme in the regeneration cycle of S-adenosylmethionine (SAM). The latter serves as a methyldonor in methylation reactions such as those required for CAP formation. Inhibition of the SAH hydrolase leads to accumulation of SAH, which serves as an inhibitor of the SAM-dependent methylation reactions, including those required for maturation of viral RNA. A variety of carbocyclic adenosine analogues are assumed to exert their antiviral action through inhibition of the SAH hydrolase. In fact, a close correlation has been detected between the antiviral effects of various carbocyclic and acyclic adenosine analogues and their inhibitory effects on cell free SAH hydrolase (Cools and De Clercq, 1989). These compounds proved to be weak inhibitors of flavivirus replication in plaque reduction assays (Neyts et al., 1996; Tseng et al., 1989) relative to the potent activity they exert against the replication of ss(−)RNA viruses such as Ebola (Bray et al., 2000; Huggins et al., 1999). As outlined previously, however, SAH hydrolase inhibitors may possibly reduce the infectivity of newly formed virus particles and have a profound effect on second and further replication cycles.
- 2021;21(7):83846.
S-adenosyl-L-homocysteine Hydrolase: Its Inhibitory Activity Against Plasmodium falciparum and Development of Malaria Drugs
Volume 2 | Issue 1 | DOI: https://doi.org/10.33696/cardiology.2.010 Predicting COVID-19 Hospitalized Patients’ Outcome with Homocysteine , Giovanni Ponti1,*,et al.
Homocysteine (Hcy), COVID-19 vulnerability, Predictor parameters, MTHFR gene, Biomarkers, MTHFR677C>T mutations .Commentary
In October 2020, we published ‘Homocysteine as a potential predictor of cardiovascular risk in patients with COVID-19’ [1]. Since then, recent scientific evidence from other authors [2-6], together with our own continuing research to include a larger cohort of hospitalized COVID-19 patients [7], has supported our original hypothesis, confirming that homocysteine (Hcy) is a predictive marker of COVID-19 outcome.
The COVID-19 pandemic has provoked a global, rapid increase of cases due to the high infectivity of the etiological agent, COVID-19 virus. In February 2021, over 110 million confirmed COVID-19 cases with 1 million deaths were reported worldwide (www.who.int).
Since the beginning of the pandemic, the identification of reliable biomarkers for COVID-19 disease progression has been a great challenge. Among various biomarkers tested, Hcy has sparked particular interest due to its association with both the metabolism of the SARS-CoV-2 virus and cardiovascular complications, which have proven to be the main cause of death among COVID-19 patients [8-10].
It is known that the SARS-CoV-2 virus transfers methyl group for viral RNA capping, from the host cell S-adenosylmethionine (SAM) converted into S-adenosylhomocysteine (SAH). SAH hydrolase (SAHH) removes adenosine from SAH, and produces an intermediate product called Hcy, “homocysteine,” which is recycled by remethylation and the trans-sulphuration pathway in the human body [5,6,11,12].
Recently, novel regulatory mechanisms directly involved with Hcy in the activation of angiotensin II type receptor have been described [13]. Ferroptosis, a newly identified form of regulated cell death, does not share morphological, biochemical, or genetic similarities with other forms of regulated cell death, such as apoptosis [14]. It is characterized by the accumulation of iron and lipid reactive oxygen species (ROS) and by smaller mitochondria with condensed membrane densities. Increasing evidence suggests that ferroptosis dysfunction is positively related to several human diseases, including tumorigenesis [15]. Ferroptosis was found to be linked to common symptoms of COVID-19 disease, namely neurological disturbances, including cognitive impairment, ageusia, and anosmia (taste and smell loss) [16].
Regarding the genetic background of Hcy metabolisms, the enzyme 5,10-methylenetetrahydrofolate reductase (MTHFR) is involved in folate metabolism. The MTHFR converts 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate, which produces methyl donor for the conversion of Hcy to methionine [17].
Hcy has been reported as a potential predictive biomarker for COVID-19 infection severity in many studies [1-4]. In a series of 273 Chinese hospitalized patients with mild COVID-19 disease, over 40 parameters were measured at admission. Disease progression was registered for 72 patients (computed tomography [CT] lung scans) and age, Hcy plasma levels and monocyte-to-lymphocyte ratio (MLR) were the only significant predictors in hyperhomocysteinemic patients (>15.4 μmol/L), estimated to correspond with a three-fold increased risk of disease evolution at radiological images. Interestingly, Hcy is the only predictive marker identified which is readily modifiable. Further, recent data confirmed the value of Hcy (together with age, MLR, and time from disease onset to hospital admission) in predicting the risk of severe pneumonia (on chest CT scan). The authors did not report any other additional organ involvement [3].
Our retrospective cohort study, including 313 COVID-19 hospitalized patients (female 34.8%; mean age 62 years) between April-September 2020, also included a broad panel of clinical laboratory data collected at admission. Of the enrolled patients, 10.9% died during hospitalization (3% were transferred to other hospitals and were lost to follow-up). Hcy was found to be the strongest predictor of Covid-19 critical-progression leading to death. Univariate analysis demonstrated that age (OR 1.04), Hcy (OR 1.06), and Neutrophil/Lympocyte count ratio (OR 1.03) were significant predictors of critical progression leading to death and RBC (OR 0.68) and Lymphocytes count (OR 0.23) with benign outcome (Table 1). ROC analysis indicated Hcy cut off of 16 μmol/L for predicting COVID-19 infection outcome (sensitivity 40% and specificity 84%); patients with Hcy levels >16 μmol/L had significantly increased risk of in-hospital mortality (p=0.002) both as a continuous and dichotomic value. Our results demonstrate that Hcy is an effective predictive biomarker for hospitalized COVID-19 patients’ outcome.
Several studies have demonstrated the importance of vitamin supplementation in patients with the COVID-19 disease. Vitamin B supplements (especially B9 and B12) are able to normalize blood Hcy levels in both apparently healthy individuals and patients with a history of stroke or Parkinson’s disease [18-20]. It is reasonable to suggest that B vitamins and Folic acid integration may have protective clinical effects for patients with infectious disease, due to MTHFR 677T allele alteration or other pathologic conditions. The relationship between the prevalence of genetic polymorphisms of MTHFR C677T and COVID-19 incidence and mortality rates seems to be intriguing; it may be useful biomarker COVID-19 infection severity stratification and it may be used for preventive medical treatments and supplementations.
Hyper-homocysteinemia is related to many virus infection outcomes, including human hepatitis virus [21], human papilloma virus [22] and Human immunodeficiency virus [23,24]. B vitamins (B2, B3 and B6) have a key role in the enhancement of the immune system [25].
Even though Hcy has been proven as a critical biomarker of cardiovascular risk and complications in hospitalized COVID-19 patients, it has not yet been adopted in prospective studies for useful laboratory markers for the stratification of COVID-19 patients.
Hcy may be a valuable biomarker which can help clinicians identify patients who are at higher risk for severe COVID-19 infection. Hcy plasma levels are easily determined by a simple and affordable laboratory test.
The association between Hcy levels >16 μmol/L and worse COVID-19 prognosis should encourage preventive health programs aimed to supplement dietary group B vitamins and folic acid for COVID-19 patients.
https://www.hsph.harvard.edu/nutritionsource/folic-acid/
Foolihapon toinen nimi on vitamiini 9 muualla maailmassa.
torsdag 11 maj 2023
B12 ja rikkiaineenvaihdunnan molekyylien osuutta metyyliryhmän siirrossa
https://pubmed.ncbi.nlm.nih.gov/28379205/
Possible Involvement of Hydrosulfide in B12-Dependent Methyl Group Transfer
- PMID: 28379205
- PMCID: PMC6154648
- DOI: 10.3390/molecules22040582
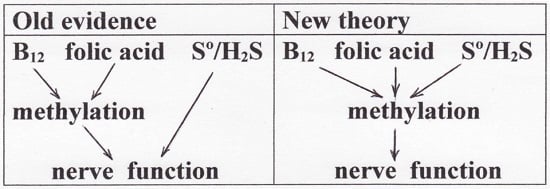
Evidence from several fields of investigation lead to the hypothesis that the sulfur atom is involved in vitamin B12-dependent methyl group transfer. To compile the evidence, it is necessary to briefly review the following fields: methylation, the new field of sulfane sulfur/hydrogen sulfide (S°/H₂S), hydrosulfide derivatives of cobalamins, autoxidation of hydrosulfide radical, radical S-adenosylmethionine methyl transfer (RSMT), and methionine synthase (MS). Then, new reaction mechanisms for B12-dependent methyl group transfer are proposed; the mechanisms are facile and overcome difficulties that existed in previously-accepted mechanisms. Finally, the theory is applied to the effect of S°/H₂S in nerve tissue involving the "hypomethylation theory" that was proposed 50 years ago to explain the neuropathology resulting from deficiency of vitamin B12 or folic acid. The conclusions are consistent with emerging evidence that sulfane sulfur/hydrogen sulfide may be beneficial in treating Alzheimer's disease.
Keywords: Alzheimer’s disease; cobalamin; dementia; hydrogen sulfide; hypomethylation; methionine auxotrophy; methionine synthase; methylation; radical S-adenosylmethionine methyl transferases; sulfane sulfur; viscose dialysis tubing.
The author declares no conflict of interest.
Desulfovibrio, Mercury , HAKU
Haku: Desulfovibrio, mecury, (amalgama)
85 articles
Mercury methylation converts inorganic mercury into the toxic methylmercury, and the consequences of this transformation are worrisome for human health and the environment. This process is performed by anaerobic microorganisms, such as several strains related to Pseudodesulfovibrio and Desulfovibrio genera. In order to provide new insights into the molecular mechanisms of mercury methylation, we performed a comparative genomic analysis on mercury methylators and non-methylators from (Pseudo)Desulfovibrio strains. Our results showed that (Pseudo)Desulfovibrio species are phylogenetically and metabolically distant and consequently, these genera should be divided into various genera. Strains able to perform methylation are affiliated with one branch of the phylogenetic tree, but, except for hgcA and hgcB genes, no other specific genetic markers were found among methylating strains. hgcA and hgcB genes can be found adjacent or separated, but proximity between those genes does not promote higher mercury methylation. In addition, close examination of the non-methylator Pseudodesulfovibrio piezophilus C1TLV30 strain, showed a syntenic structure that suggests a recombination event and may have led to hgcB depletion. The genomic analyses identify also arsR gene coding for a putative regulator upstream hgcA. Both genes are cotranscribed suggesting a role of ArsR in hgcA expression and probably a role in mercury methylation.
Keywords: Comparative genomics; Phylogeny; Regulation; Sulfate reducing bacteria; Synteny.